Add annotation
1 General
- Type: - Matrix Processing
- Heading: - Annot. columns
- Source code: AddAnnotationToMatrix.cs
2 Brief description
Based on a column containing protein (or gene or transcript) identifiers this activity adds columns with annotations. These are read from specifically formatted files contained in the folder conf/annotations in your Perseus installation. Species-specific annotation files generated from UniProt can be downloaded from the link specified in the menu at the blue box in the upper left corner.
3 Parameters
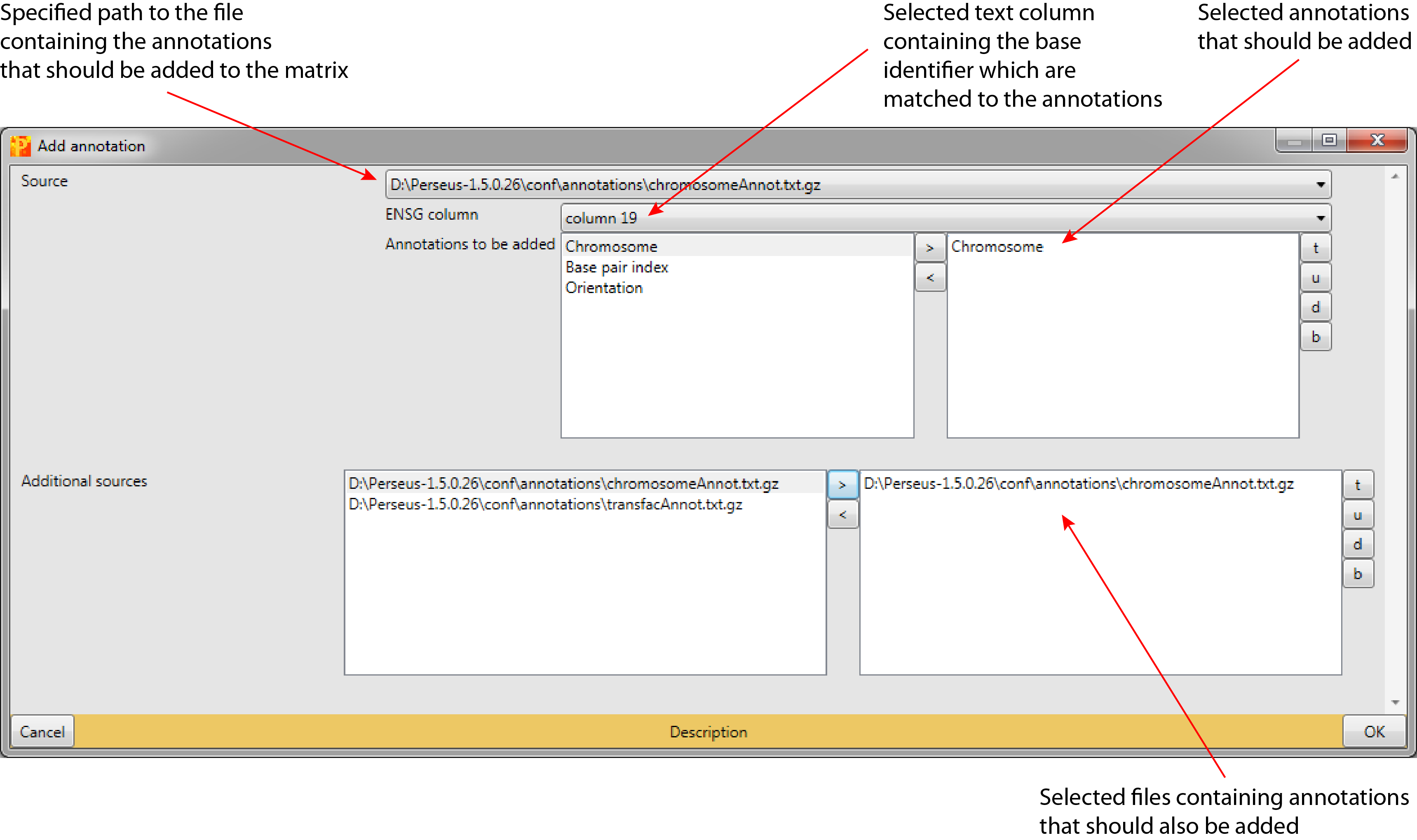
3.1 Source
Specified path to the file containing the annotations that should be added to the matrix (default: first file in Perseus-version /conf/annotations/). The file can be selected from all files in Perseus-version /conf/annotations/.
3.1.1 ENSG column
Selected text column that contains the base identifier, which are going to be matched to the annotations (default: first text column in the matrix).
3.1.2 Annotations to be added
Selected annotations that should be added (default: no annotations are selected). The annotations can be selected from a list of annotations defined by the tab separated columns in the file specified in “Source”.
3.2 Additional sources
Selected files containing the annotations that should additionally be added to the matrix (default: no files are selected). Additional sources can be selected from all files in Perseus-version conf/annotations.
4 Parameter window