Andromeda Enzymes - the proteases table
1 Configure the proteases table
For the here shown step by step description Andromeda was used within MaxQuant (version 1.5.3.8).
1.1 Open the proteases table
Open MaxQuant and go to the Andromeda configuration tab. There select the Data type “Proteases” (s. Figure 1).
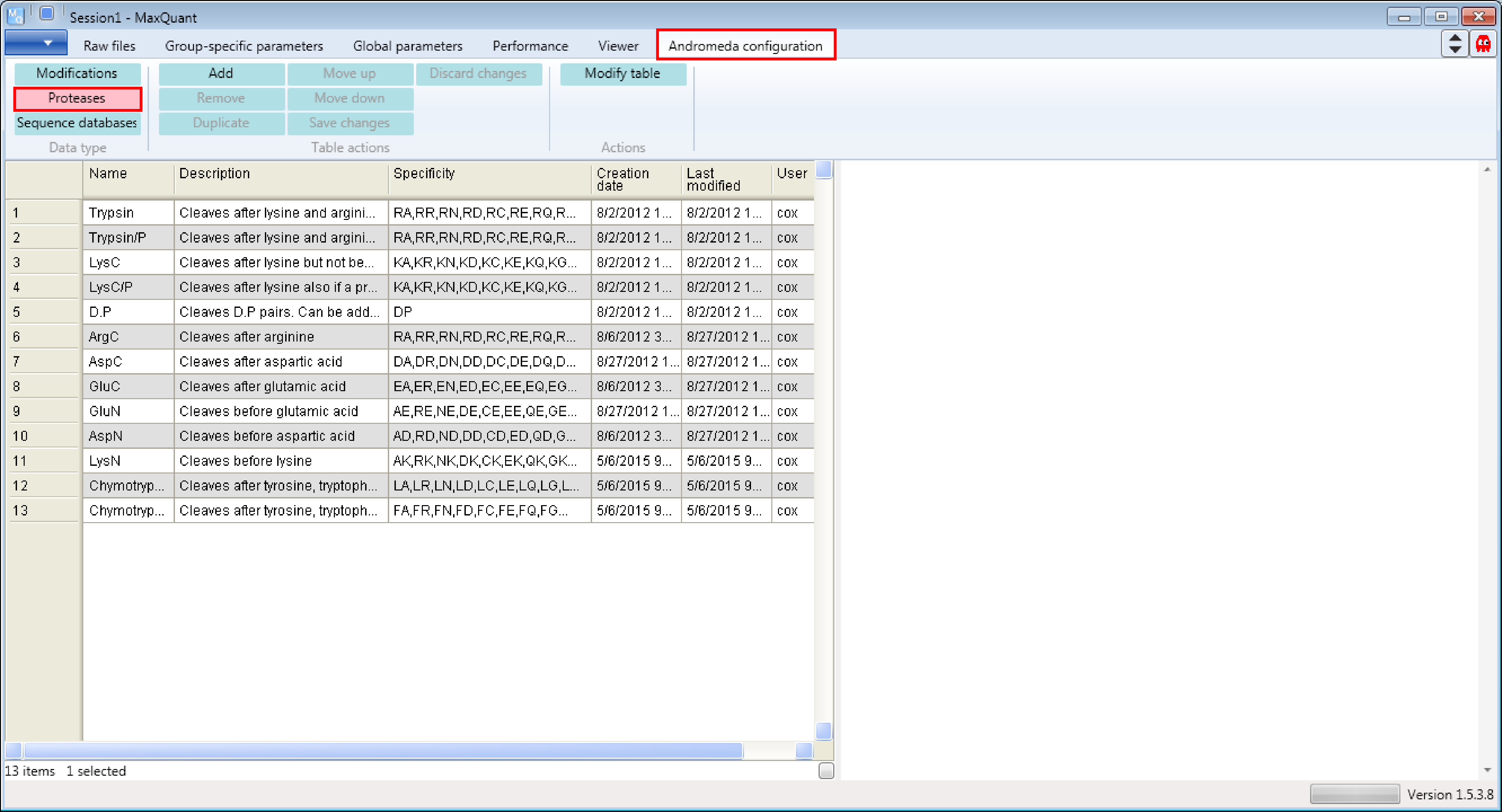
1.2 Viewing examples
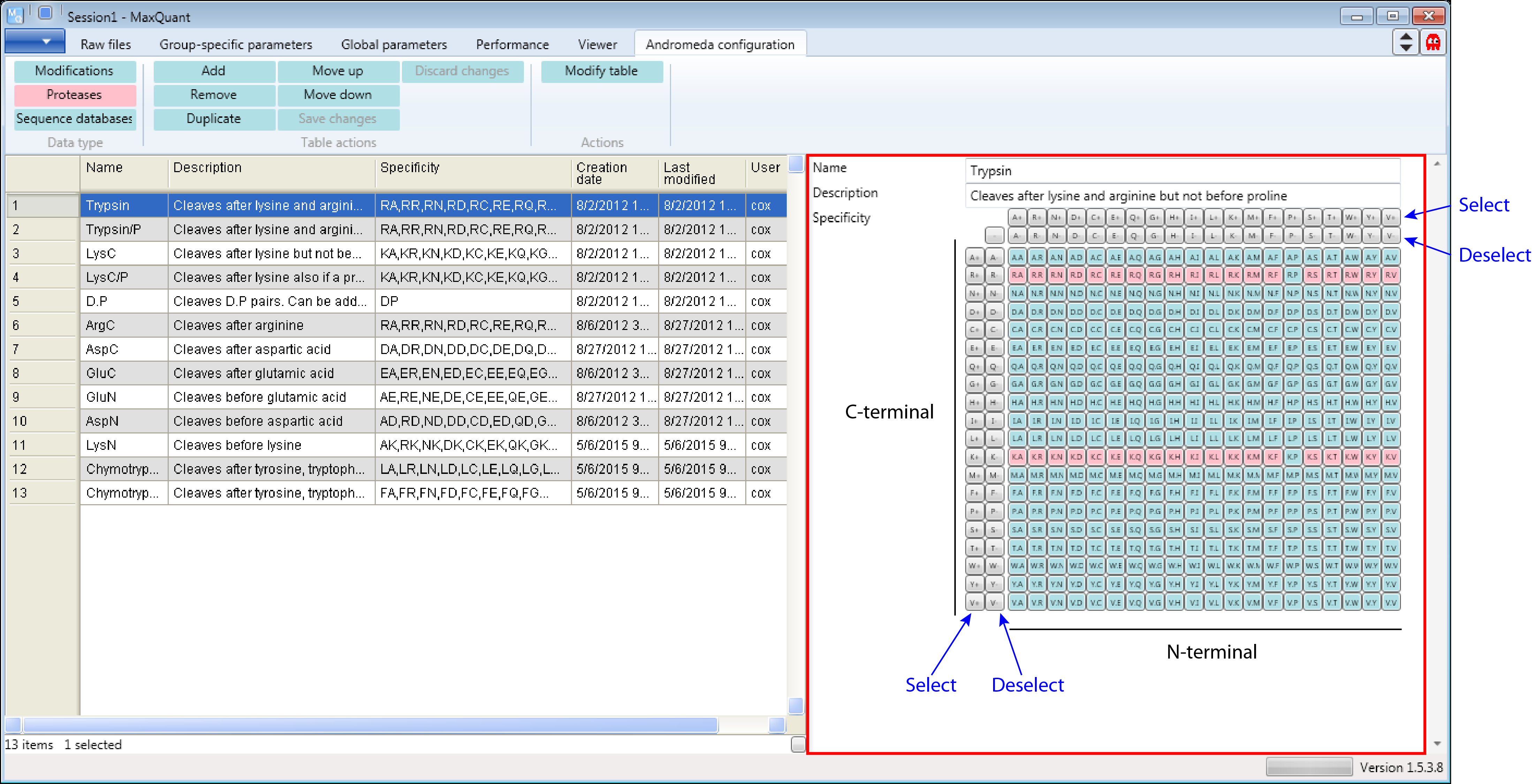
In most studies samples are digested using Trypsin. In the “Proteases” table you will find two different definitions for Trypsin. The first definition cleaves at the carboxyl side of the amino acids lysine or arginine, except when either is followed by proline (see Description on the right hand side of the Andromeda window). That’s the classical definition. Additional comments in Figure 2 are in black and blue.
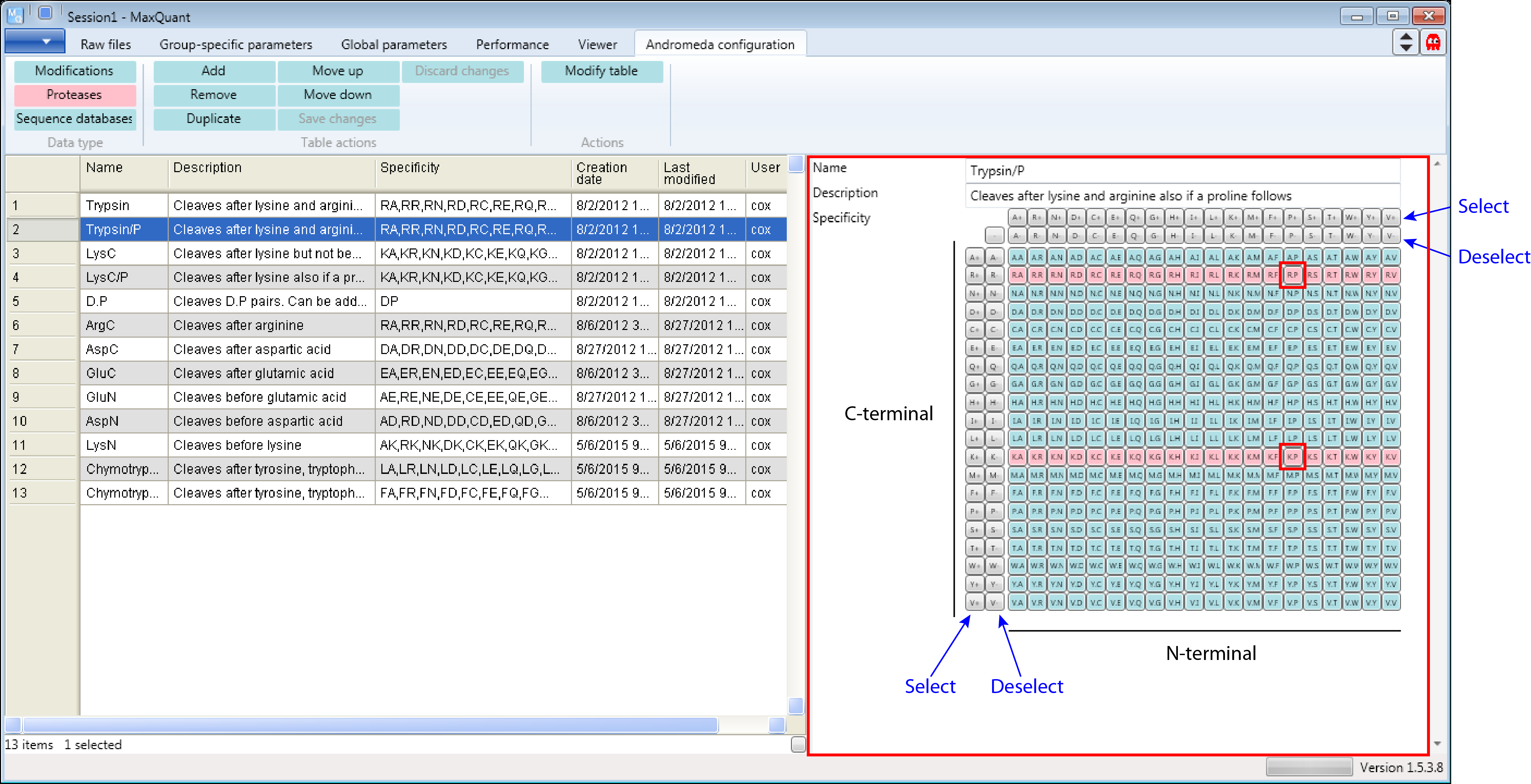
However the commonly used definition is “Trypsin/P”, which also cleaves at carboxyl side of the amino acids lysine or arginine, also if a proline follows (highlighted in red in Figure 3). Additional comments in Figure 3 are in black and blue.
1.3 Adding a new protease
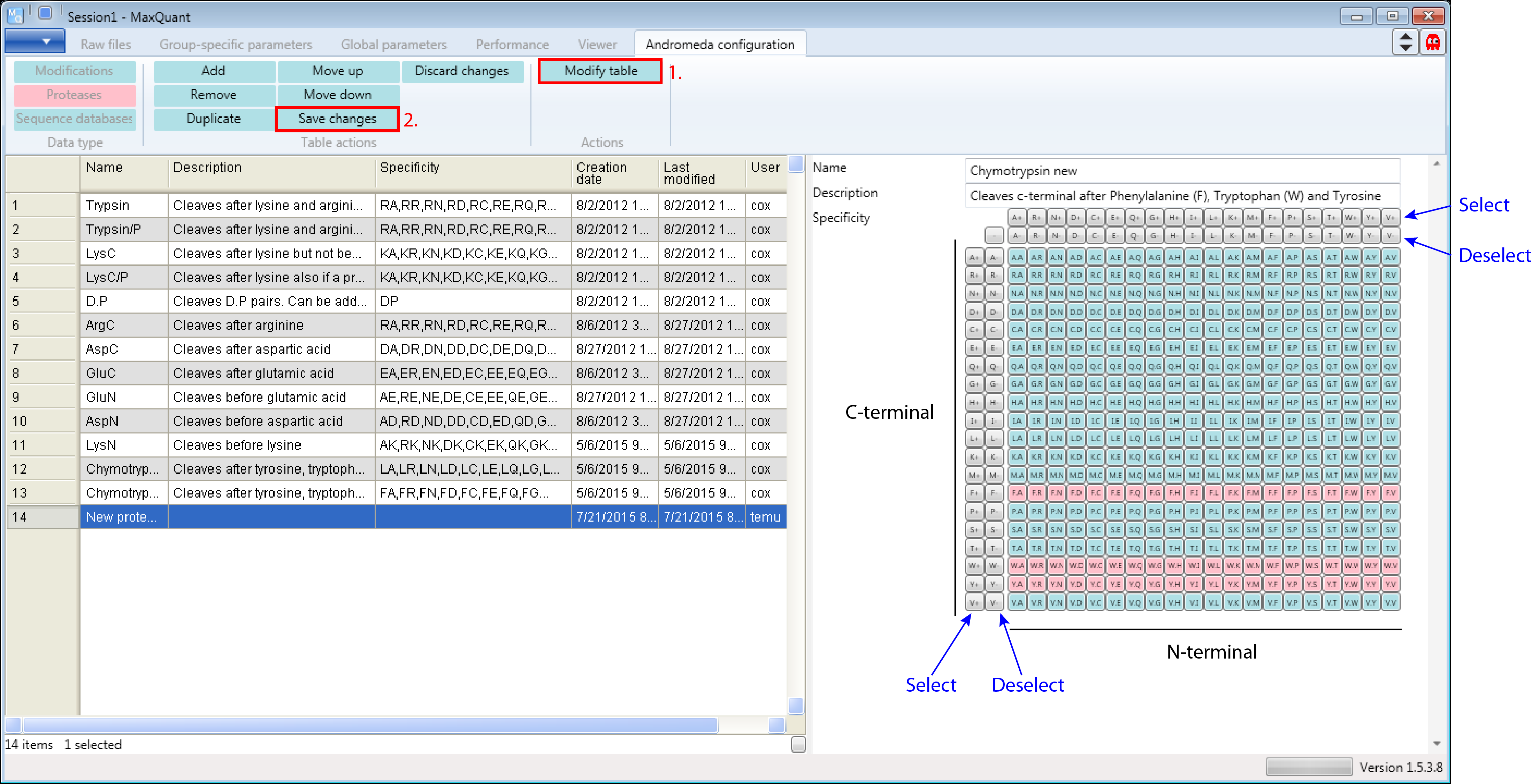
Let’s assume Chymotrypsin is not yet provided in Andromeda and we want to add it. From the literature we know Chymotrypsin cleaves c-terminal after Phenylalanine (F), Tryptophan (W) and Tyrosine (Y).
First click the “Add” button as shown in Figure 4. Then a new row will be added at the end of the table and a new protease form will appear on the right hand side that can be edited.
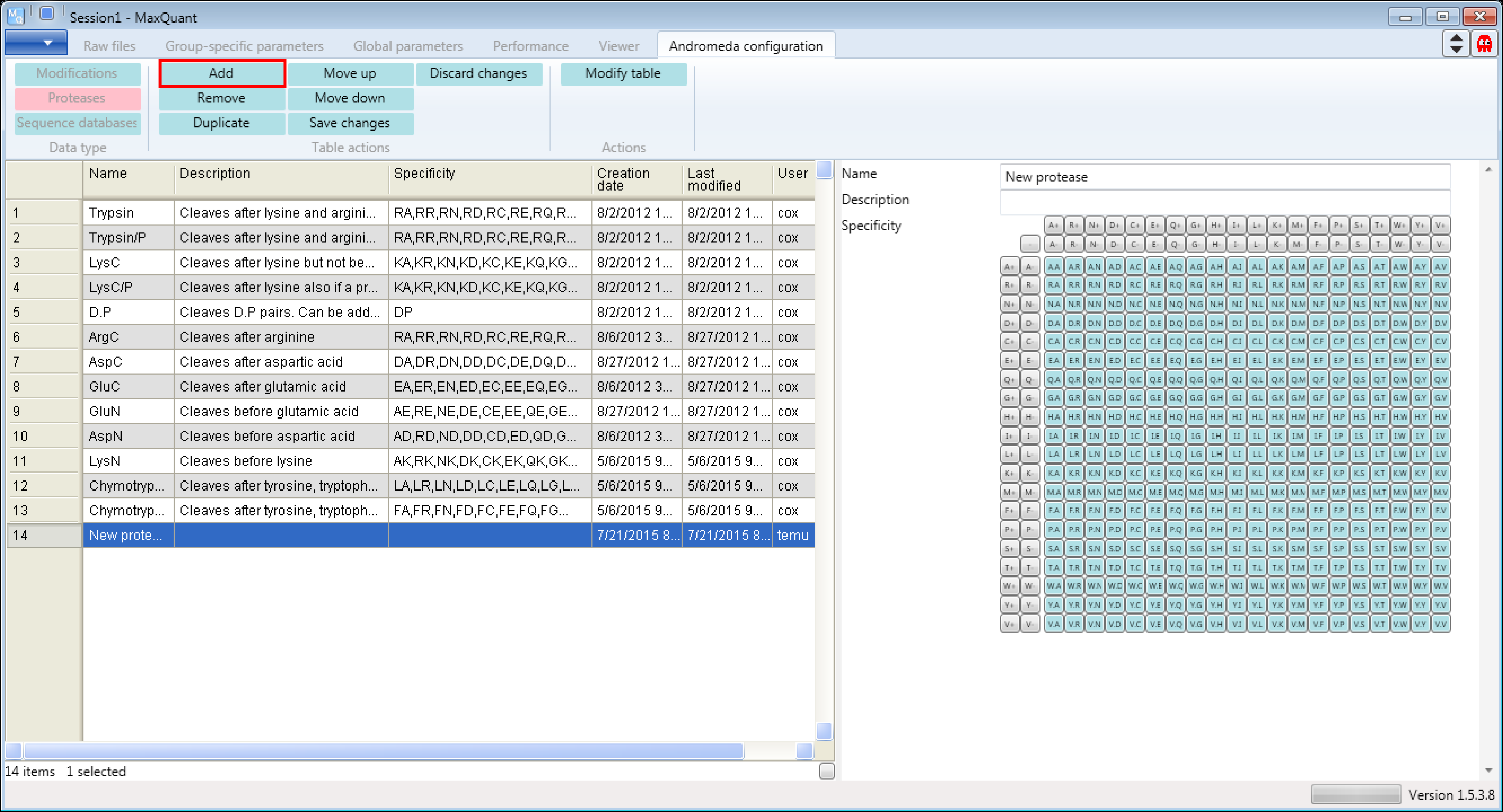
Then you just have to fill in the form by defining a name, a description and the specificity. Don’t forget to click the “Modify table” button when your done to transfer the changes you made in the form to the table on the left. And to save the table you have to click the “Save changes” button. Additional comments on the screenshot are in black and blue in Figure 5.
To have the added modification available in MaxQuant you have to open a new MaxQuant window.
Comment: Note that you also can do completely unspecific searches in MaxQuant. For this no definition of an enzyme is necessary.