Andromeda modifications table
1 Configure the modifications table
For the here shown step by step description Andromeda was used within MaxQuant (version 1.5.3.8).
1.1 Open the modifications table
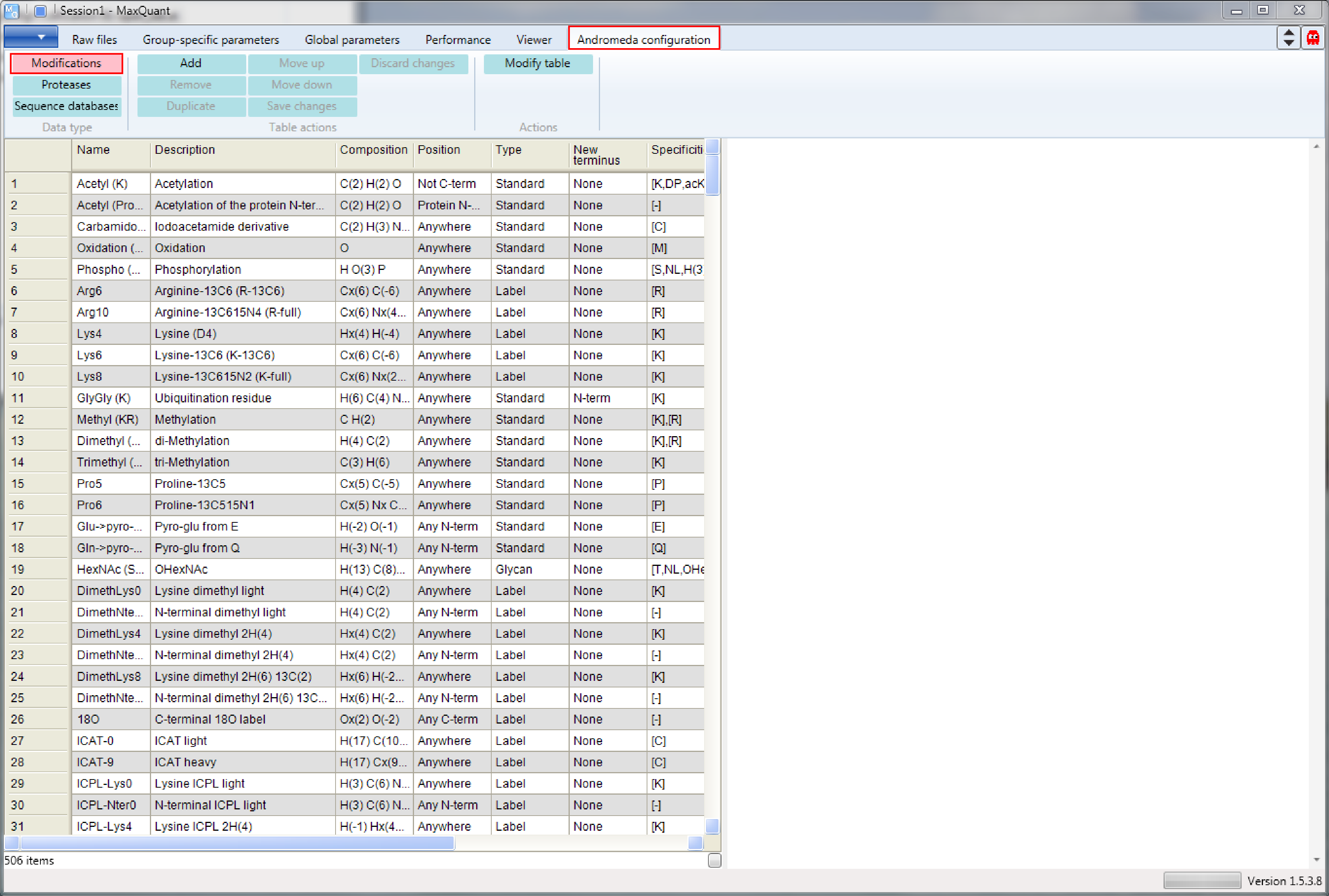
Open MaxQuant and go to the Andromeda configuration tab. The “Modifications” page is already selected Figure 1.
1.1.1 Viewing an example
First we will have a look at a modification to understand how modifications are defined in MaxQuant.
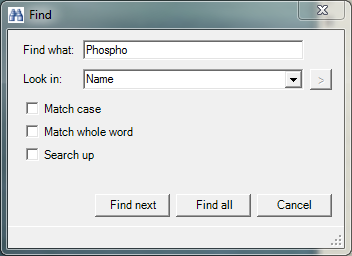
Choose the “Phospho (STY)” modification in the table on the left hand side. You can either browse through the table or use the "Find..." function of Andromeda by right clicking on the table and choosing it from the appearing menu. Then a pop-up window will show up and you can specify what you are searching and whether the term should be searched (parameter “Look in”) in the whole table or in a specific column. in Figure 2 we search in the “Name” column.
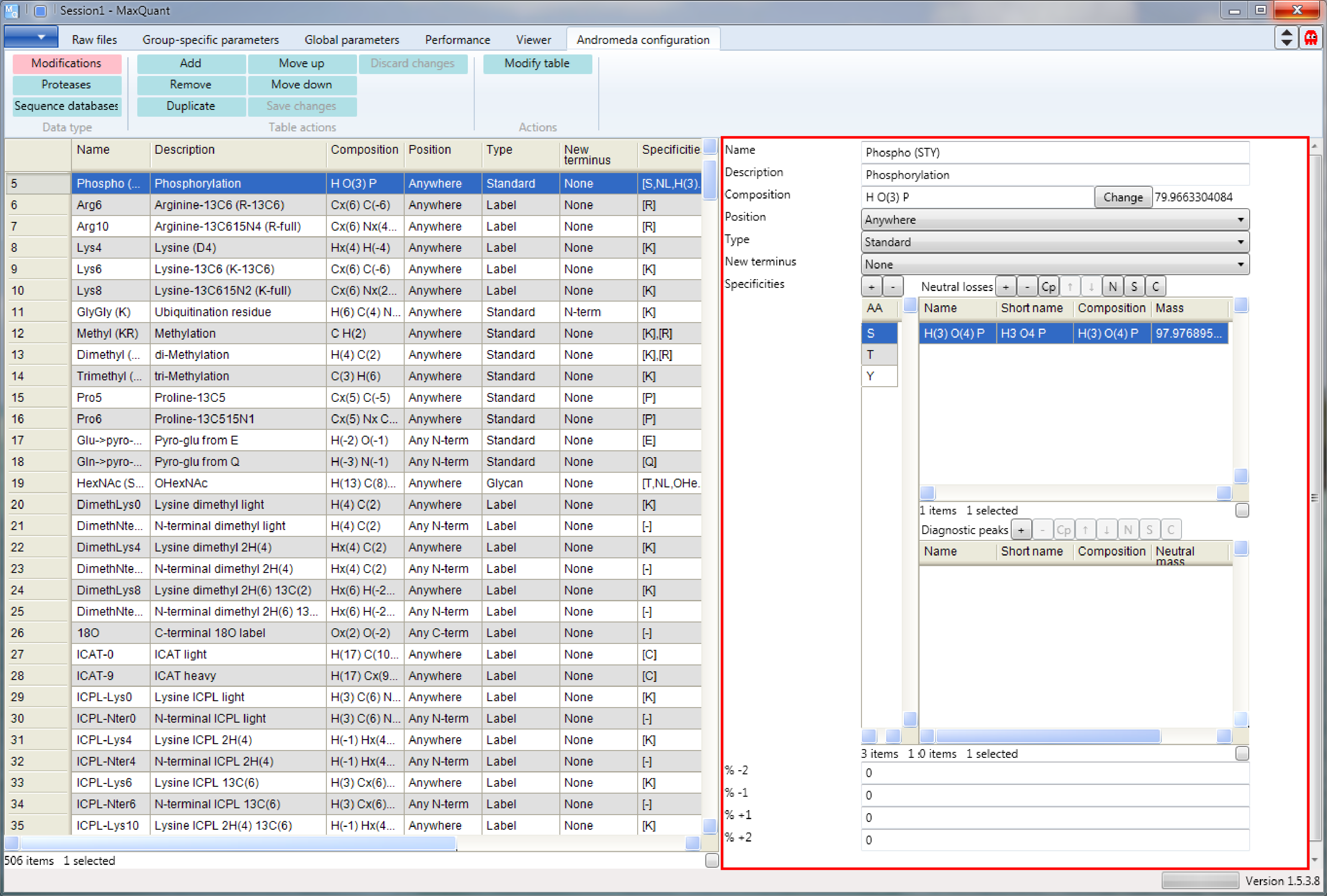
The row will be selected and the settings of the modification appear on the right hand side of the Andromeda window (s. Figure 3. Only one modification is defined together for S, T and Y. This is important for the proper calculation of localization scores for phosphorylation sites. You will notice that a neutral loss of a phosphate is only defined for serine and threonine by clicking on the amino acids in the window on the right.
A diagnostic peak of composition \(C_8H_{10}O_4P\) is only defined for tyrosine, which is the characteristic immonium ion for tyrosine phosphorylation. This will only be relevant for fragmentation spectra in which the lower mass range is available.
1.1.2 Adding a new modification
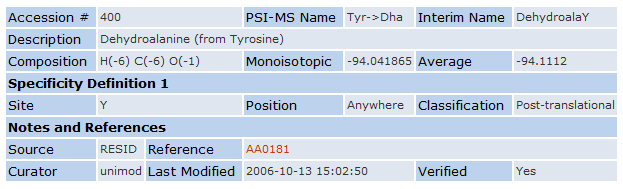
Now, we will add a new Modification. You can visit unimod.org and select a modification that has not been defined yet in MaxQuant and add this modification to the modification list in MaxQuant. Here we are adding Dehydroalanine from Tyrosine. All the information that we need can be extracted from the unimod homepage (Figure 4) like the compositional change as well as the site specificity.
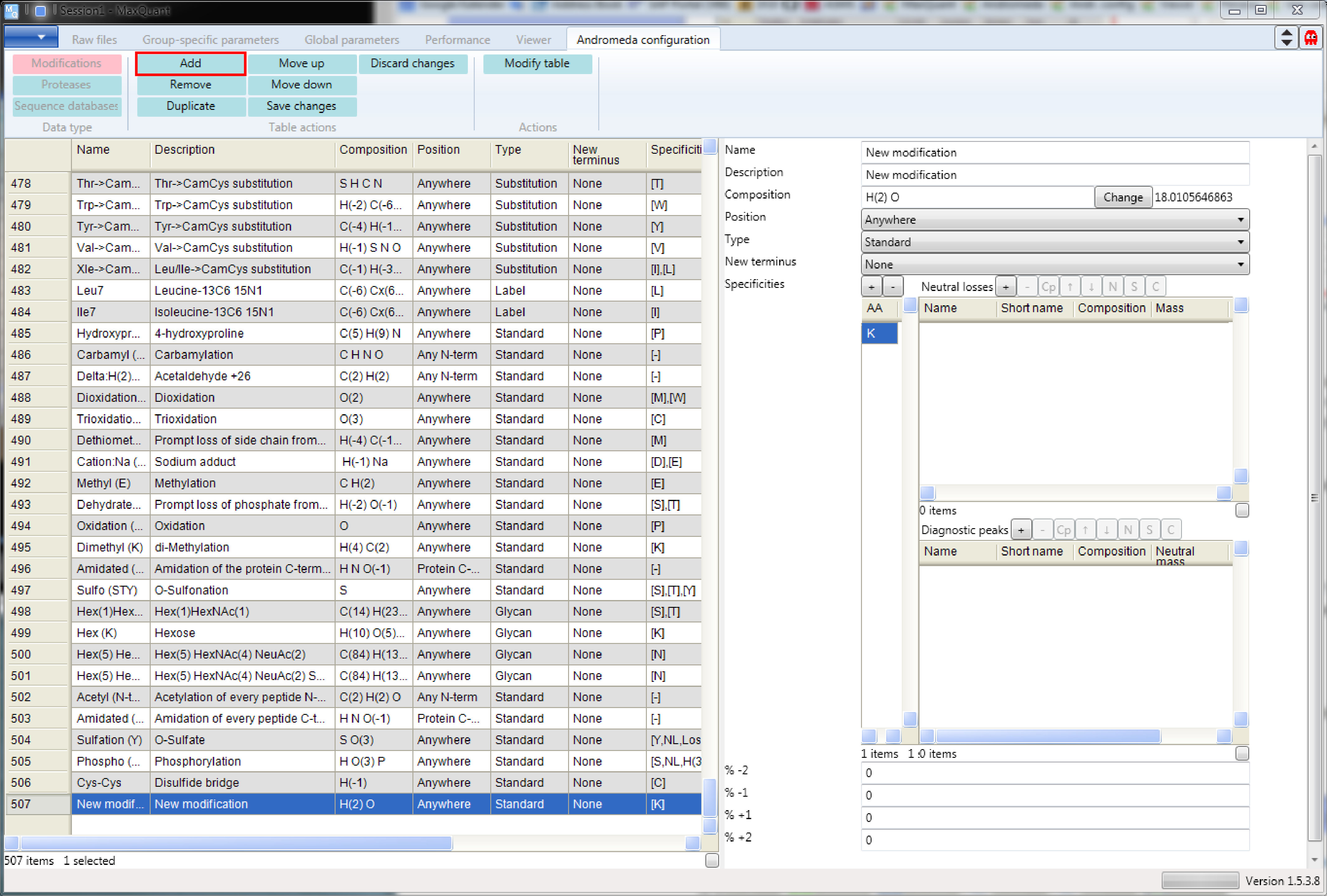
First click the “Add” button (left side of the ribbon on Figure 5). Then a new row will be added at the end of the table and a new modification form will appear on the right hand side that can be edited.
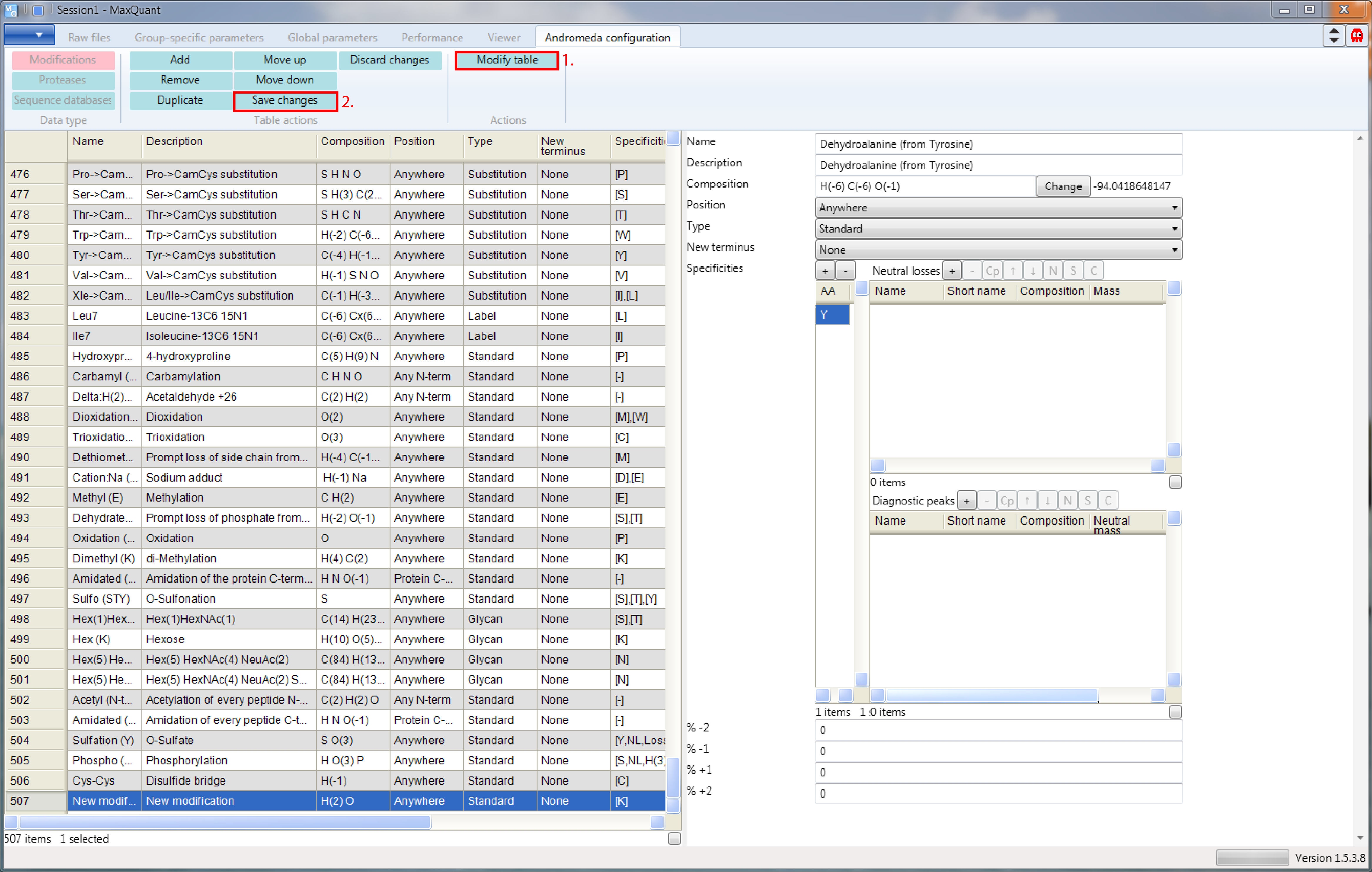
Then you just have to transfer the information from the unimod table to the form. Note that when your done you have to press the “Modify table” button to transfer the changes you made on the right side to the table on the left. And to save the table to the modifications.xml file you press the button “Save changes” (highlighted in Figure 6.
To have the added modification available in MaxQuant you have to open a new MaxQuant window.
1.1.3 Adding a new label
Note that the isotopic and isobaric labels are also defined in this table. Labels have written “Label” in the “Type” column and are the labels that can be used in MaxQuant for MS1 level quantification. Isobaric labels can also be identified by their term “isobaric Label” in the “Type” column.
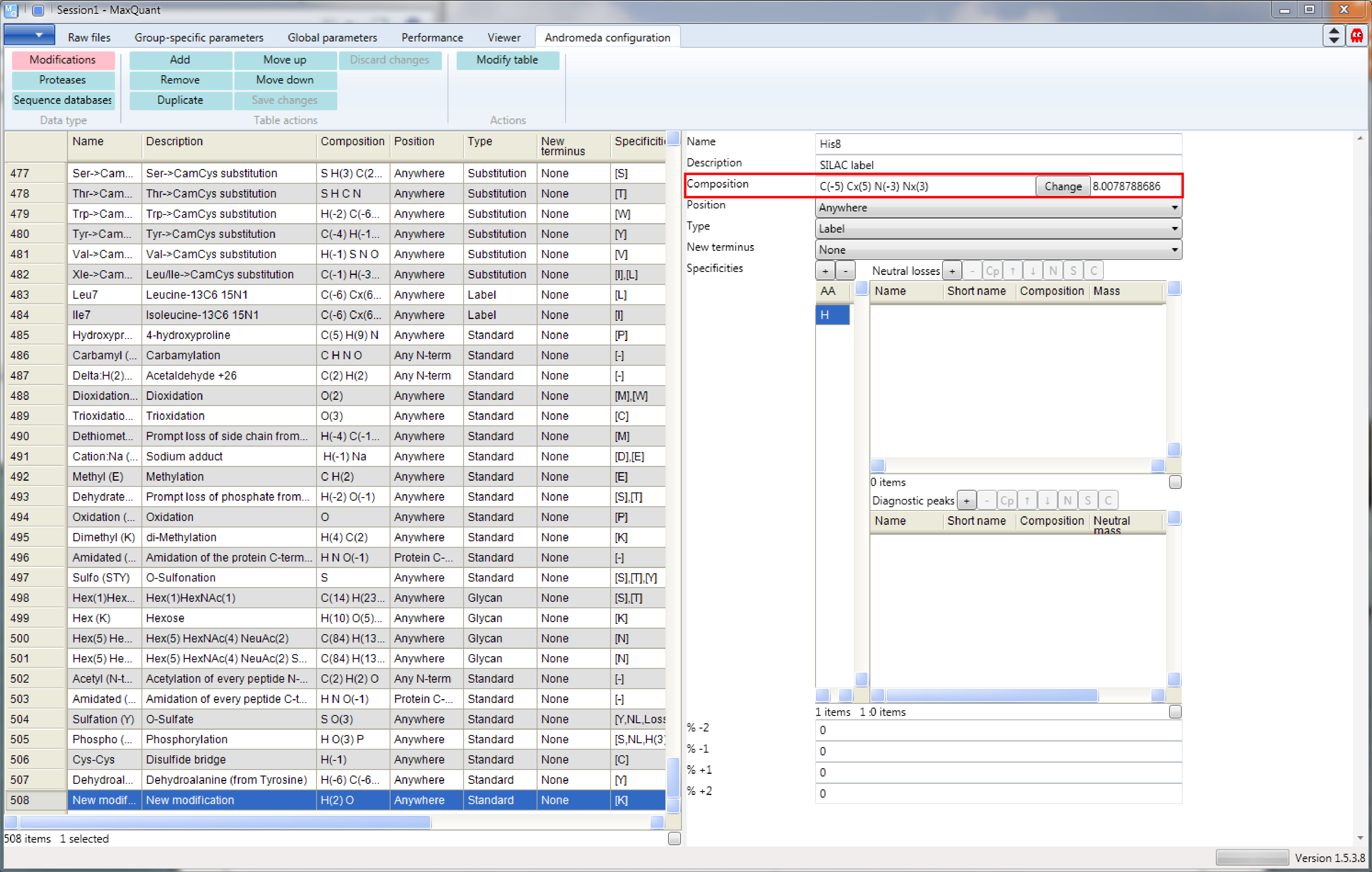
Now we define a new hypothetical SILAC label, His8 (\(His^{13}C_5^{15}N_3\)). (s. Figure 7) Therefore, we add a new entry to the modifications table.

Then the form on the right can be edited. It is important to keep in mind that we are not just adding heavy amino acids, but also have to remove the same amount of “normal” amino acids. Don’t forget to press the “Modify table” button when your done to transfer the changes you made on the right side to the table on the left. And to save the table to the modifications.xml file you have to click the “Save changes” button.
To have the added modification available in MaxQuant you have to open a new MaxQuant window.