Andromeda Protdatabases - the (protein) sequence databases table
1 Configure the sequence databases table
For the here shown step by step description Andromeda was used within MaxQuant (version 1.5.3.8).
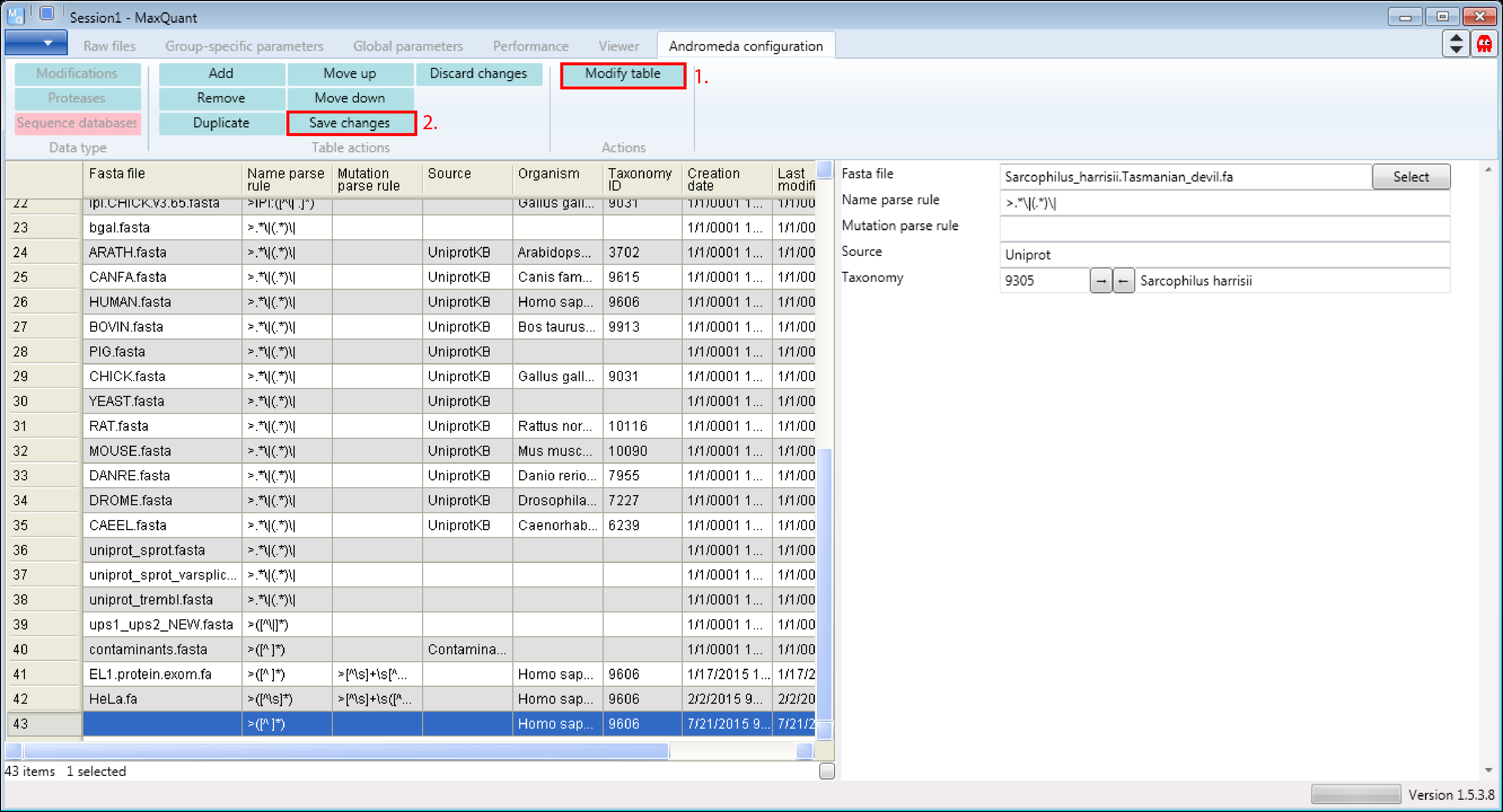
1.1 Open the sequence databases table
Open MaxQuant and go to the Andromeda configuration tab. There select the Data type “Sequence databases” (s. Figure 1).
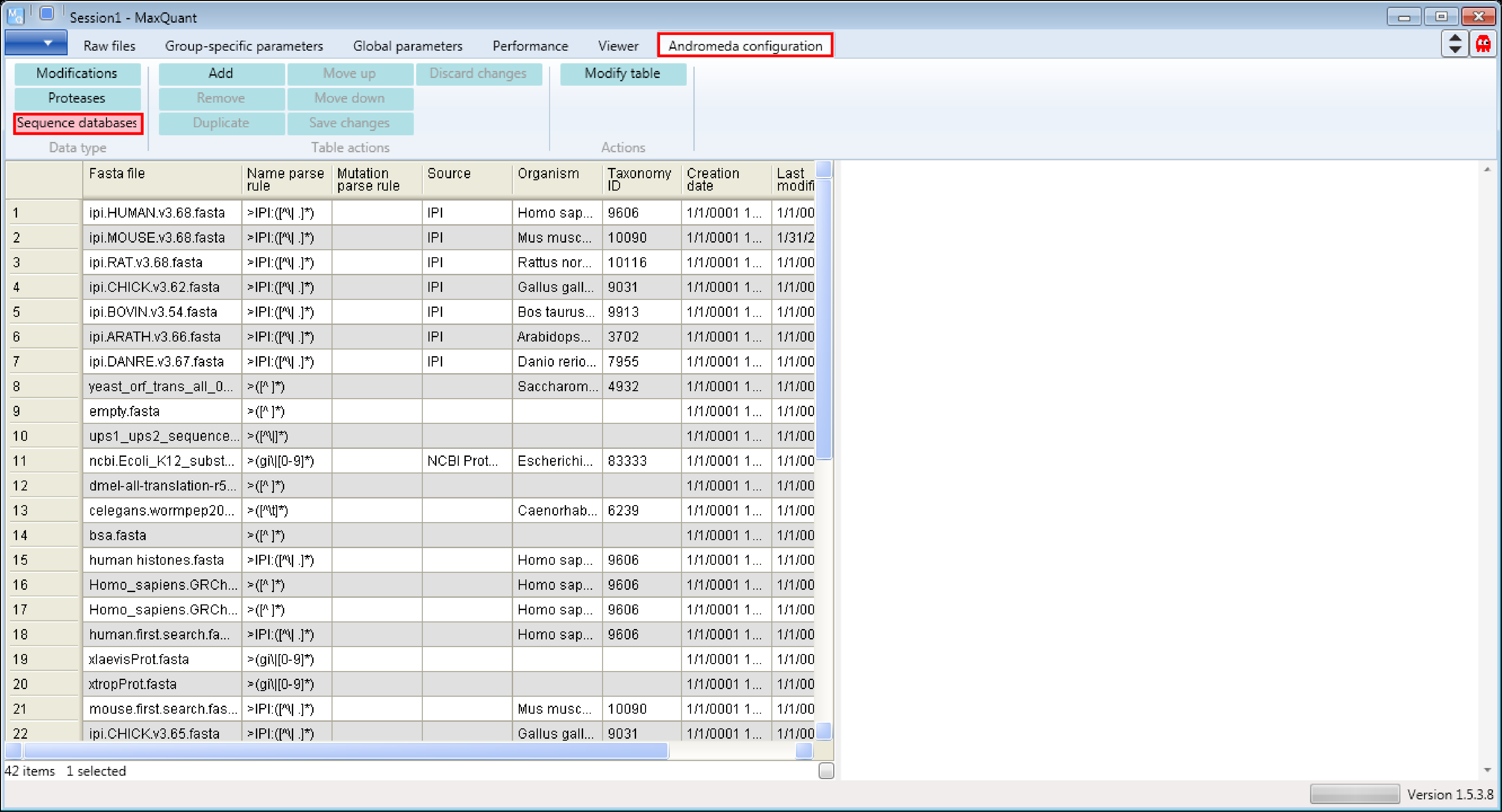
1.2 Viewing an example
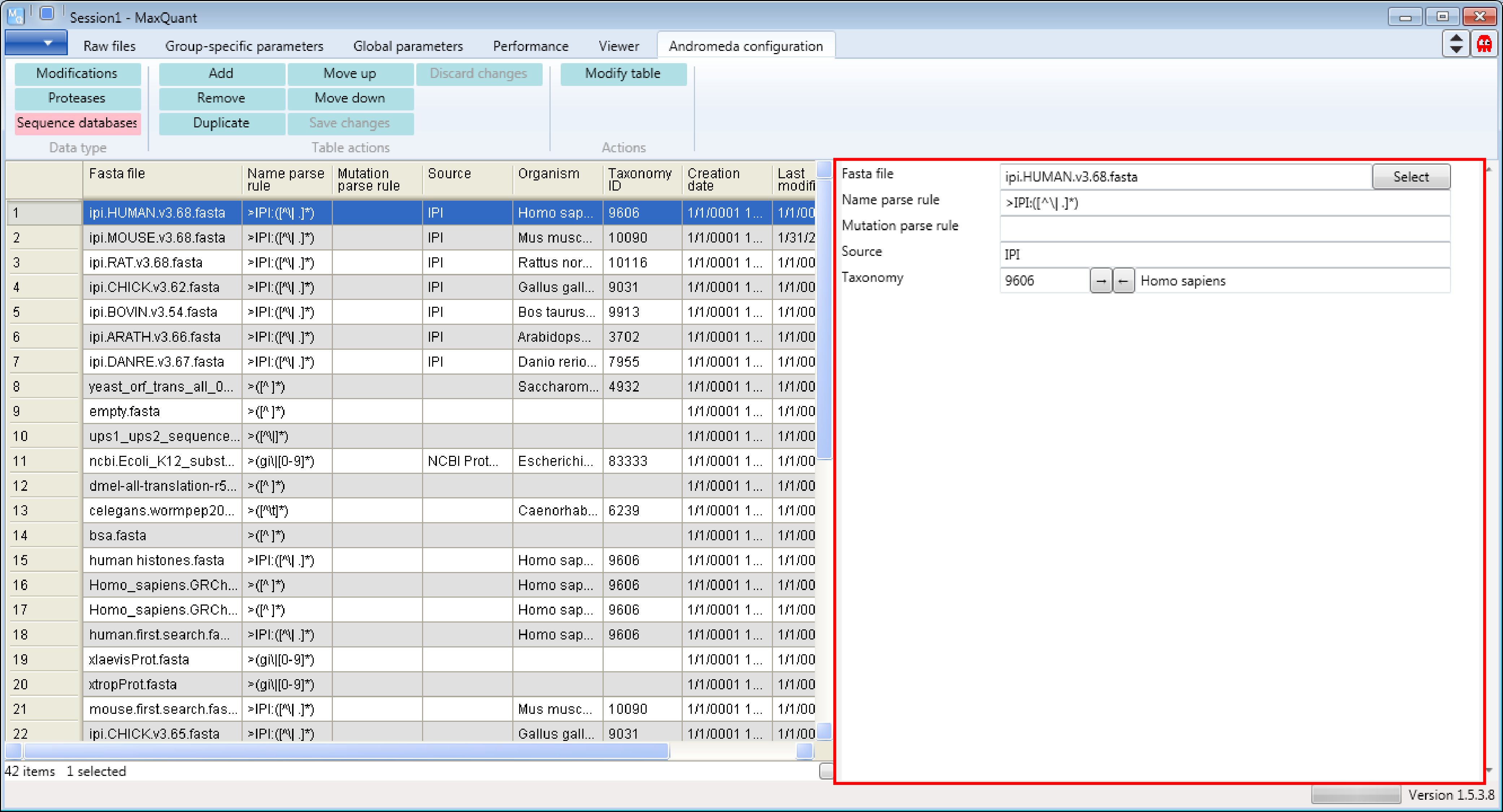
Let’s assume we have to analyze human proteome measurements and we are interested which human database is used and how it is used (see Description on the right hand side of the Andromeda window in Figure 2).
1.3 Adding a new database
Now we are assuming that a reviewer asks us to search our human proteome measurements against the tasmanian devil proteome ( Sarcophilus harrisii ) - for some reason - and we downloaded the corresponding fasta file from Uniprot.
After downloading the file, we are including the database into MaxQuant. First, click the “Add” button (highlithed in Figure 3). Then a new row will be added at the end of the table and a new sequence database form will appear on the right hand side that can be edited.
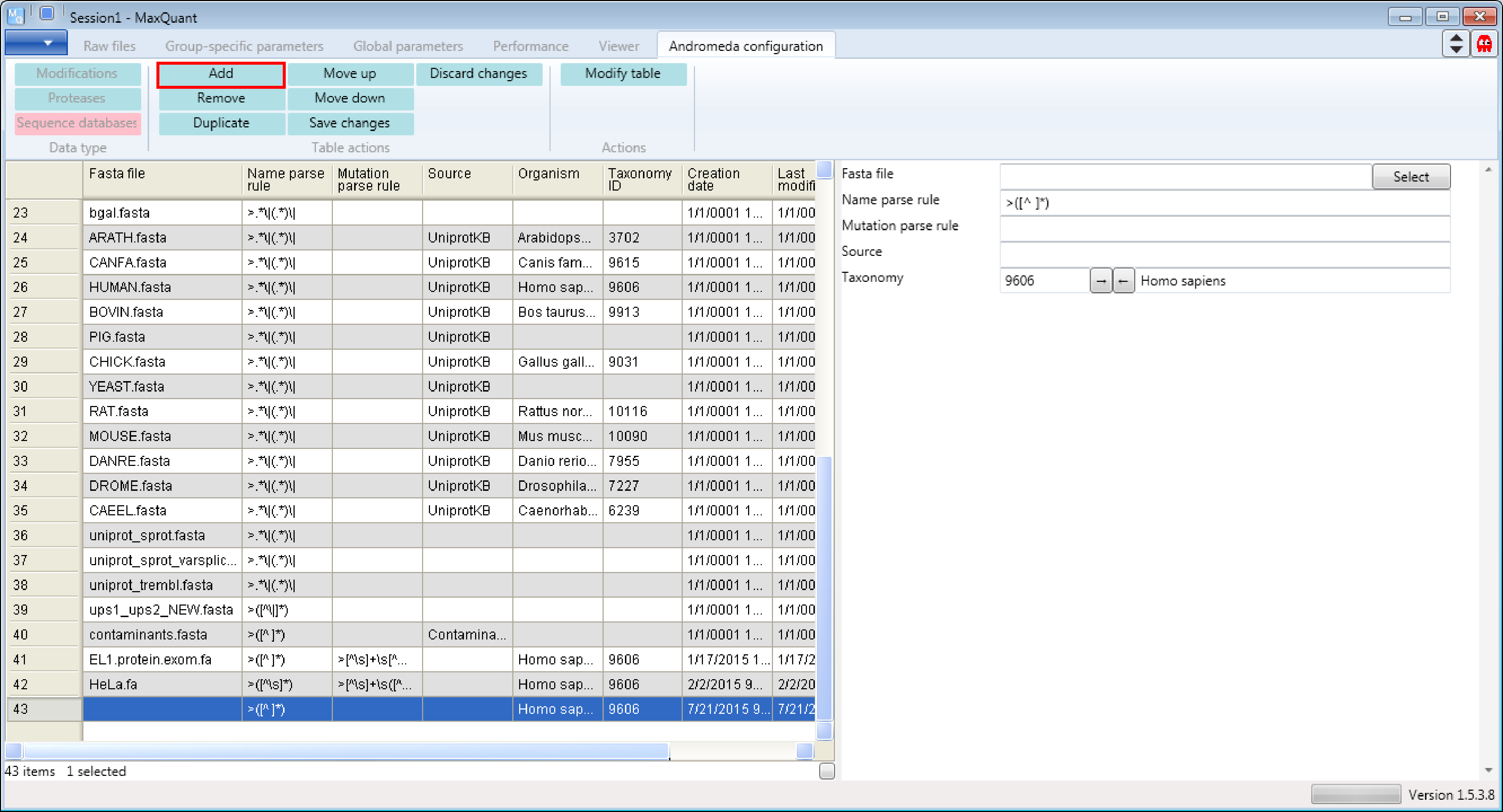
Then you just have to fill in the form by defining the fasta file that should be used, the parsing rule that should be applied to retrieve the identifiers, the source of the fasta file, the taxonomy and the organism name. To define the parsing rule regular expressions are used. A regular expression is a sequence of characters that forms a search pattern with a special syntax. A good general introduction can be found, as always, on Wikipedia. If you already know generally how regular expressions work, you may only need to glance at a this Quick Reference or at an even quicker Cheat Sheet. Also you don’t have to know the taxonomy of your organism, just type in the name and use the arrows in the taxonomy line and MaxQuant will complete the form.
Don’t forget to click the “Modify table” button (Figure 4) when you’re done to transfer the changes you made in the form to the table on the left. And to save the table you have to click the “Save changes” button.
To have the added sequence database available in MaxQuant you have to open a new MaxQuant window.