Fisher exact test
1 General
- Type: - Matrix Processing
- Heading: - Annot. columns
- Source code: not public.
2 Brief description
This is a test used to determine if there are non-random associations between a categorical column and all terms in the other categorical columns. Note that significantly enriched as well as depleted terms will be reported. Depleted terms will have an enrichment factor < 1.
Output: A new table is generated in which each row corresponds to a term in a categorical column that is significantly associated with a term in the selected categorical column.
3 Parameters
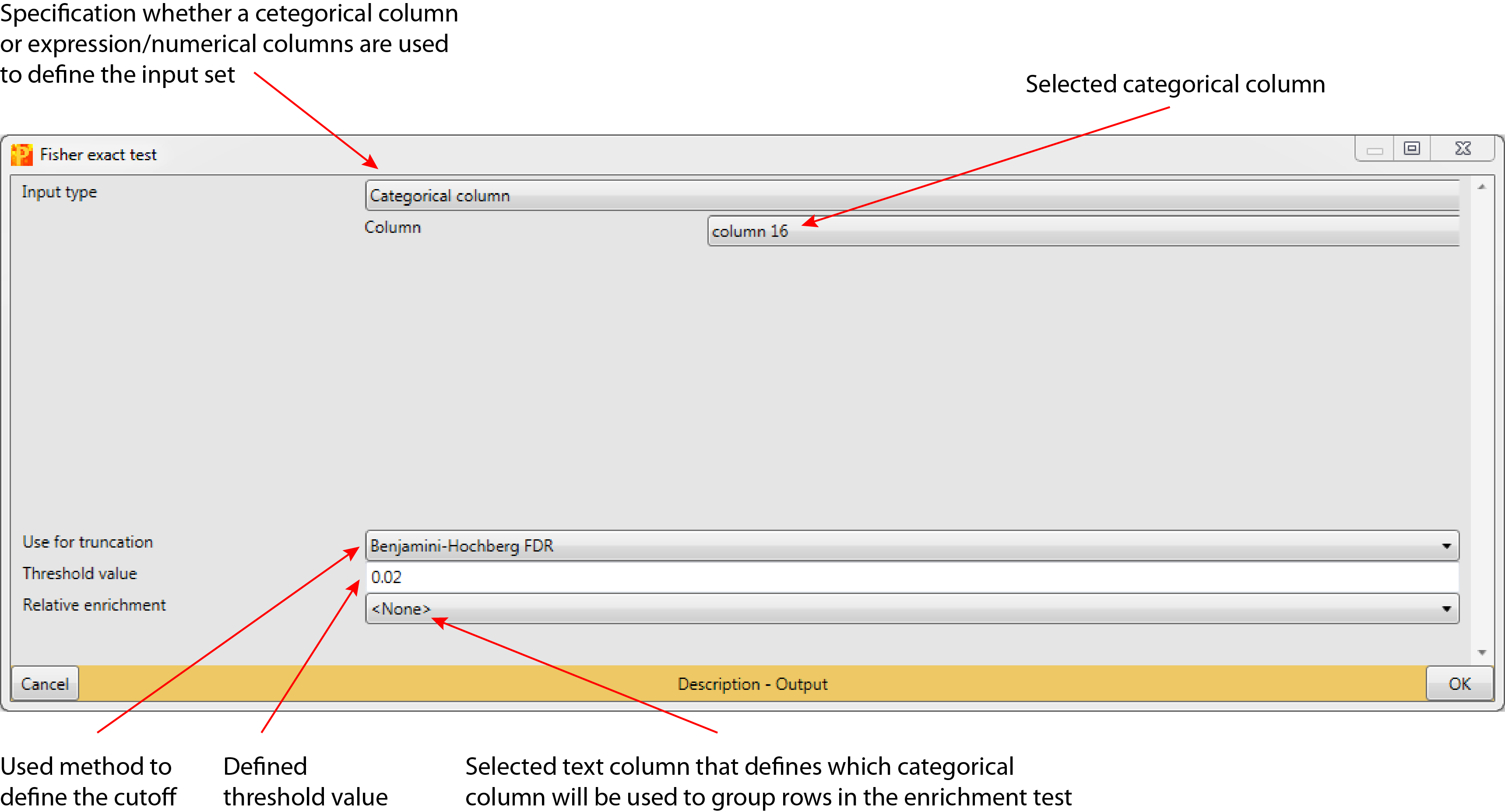
3.1 Input type
Specification whether the sub-population that is tested for contingency against all other categorical columns is taken directly from a categorical column (“Categorical column” is default) or, if it is defined by a threshold on a numerical/expression column (“Numerical column”).
3.1.1 Column
This parameter is just relevant, if “Input type” is set to “Categorical column”. The selected categorical column is checked against all other categorical columns for association between the occurrence of terms (default: first categorical column in the matrix).
3.1.2 Columns
This parameter is just relevant, if “Input type” is set to “Numerical column”. The selected expression/numerical columns are used as a threshold to define the set of interest. The set is then checked against all categorical columns for association between the occurrence of terms (default: all expression and numerical columns are selected).
3.1.3 Threshold
This parameter is just relevant, if “Input type” is set to “Numerical column”. It defines the threshold, which rows are kept/discarded to define the set of interest (default: 2).
3.1.4 Selection is
This parameter is just relevant, if “Input type” is set to “Numerical column”. It defines, whether the values in the selected “Columns” should be “Larger than threshold” or “Less than threshold” (default: Larger than threshold).
3.2 Use for truncation
The truncation can be based on p-values or the Benjamini-Hochberg correction for multiple hypothesis testing (default: Benjamini-Hochberg, FDR). Rows with a test result below a specified value (Section 3.3) are reported as significant.
3.3 Threshold value
Based on a specified threshold (default: 0.02) a specific row is reported as significant. Depending on the chosen truncation score (Section 3.2) this threshold value is applied to the p-value or to the Benjamini-Hochberg FDR.
3.4 Relative enrichment
Selected text column, where all rows having the same identifier will be counted as one entity in the Fisher exact test (default:
4 Parameter window