Multiple-samples tests
1 General
- Type: - Matrix Processing
- Heading: - Tests
- Source code: not public.
2 Brief description
Multi-sample test for determining if any of the means of several groups is significantly different from each other.
Output: A numerical columns is added containing the p-value. In addition there is a categorical column added in which it is indicated by a ‘+’ when the row is significant with respect to the specified criteria.
3 Parameters
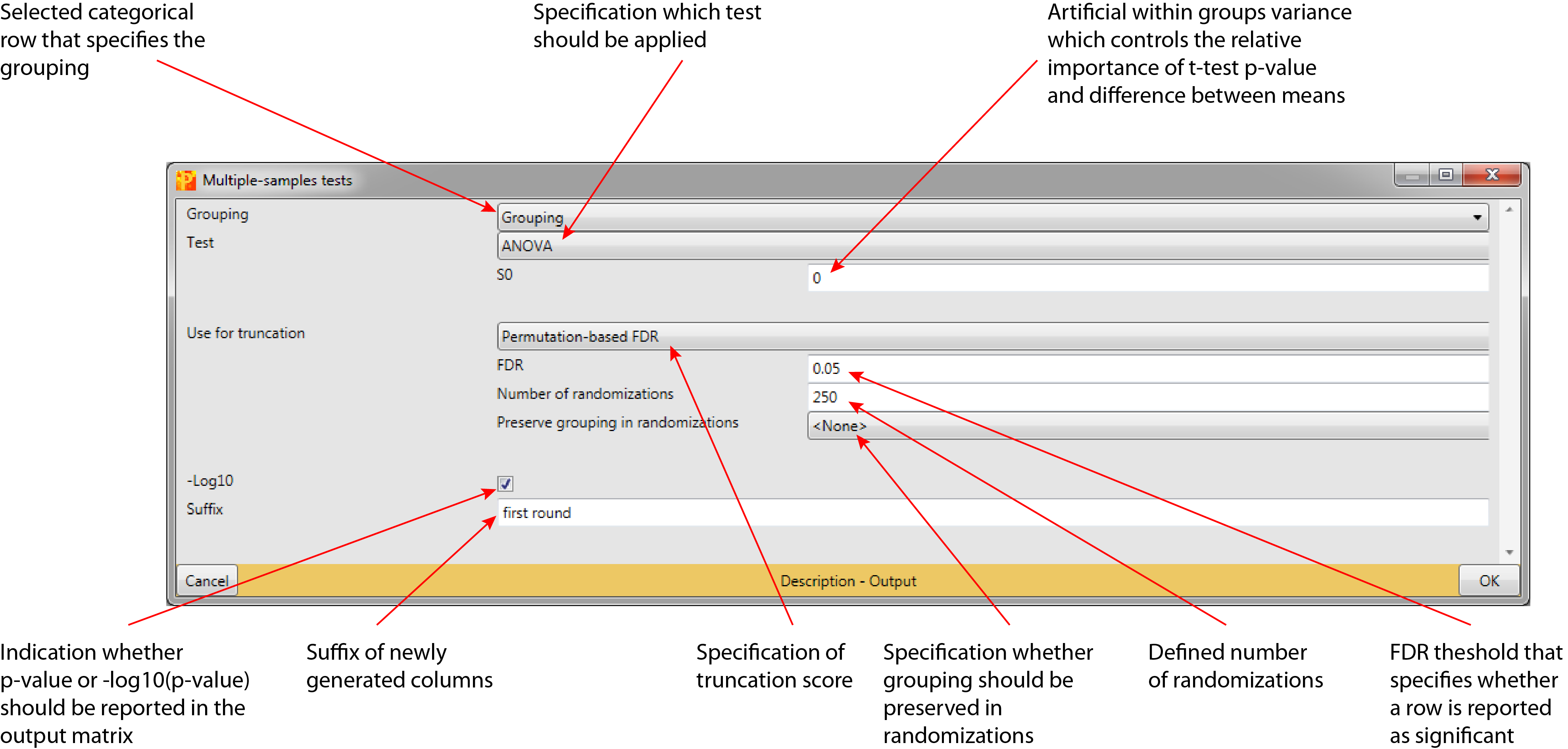
3.1 Grouping
Selected categorical row that defines the grouping of columns that should be used in the test (default: first categorical row in the matrix).
3.2 Test
Defines what kind of test should be applied (default: ANOVA). The test can be selected from a predefined list:
- ANOVA
- Kruskal Wallis
3.2.1 S0
Artificial within groups variance (default: 0). It controls the relative importance of t-test p-value and difference between means. At \(s0=0\) only the p-value matters, while at nonzero s0 also the difference of means plays a role. See (Tusher, Tibshirani, and Chu 2001) for details.
3.3 Use for truncation
Defines on what value the truncation is based on (default: Permutation-based FDR). Choose here whether the truncation should be based on the p-values, on permutation-based FDR-values or, if the Benjamini-Hochberg correction for multiple hypothesis testing should be applied.
3.3.1 Threshold p-value
This parameter is just relevant, if the parameter “Use for truncation” is set to “P-value”. Rows with a test result below this value are reported as significant (default: 0.05).
3.3.2 FDR
This parameter is just relevant, if the parameter “Use for truncation” is set to “Benjamini-Hochberg FDR” or “Permutation-based FDR”. Rows with a test result below this value are reported as significant (default: 0.05).
3.3.3 Number of randomizations
Specifies the number of randomizations that should be applied (default: 250).
3.3.4 Preserve grouping in randomizations
Defines, whether the grouping specified in a categorical row should be preserved in the randomizations (default: <None>). It can be selected from a list including all available groupings of the matrix.
3.4 Log10
If checked, \(-Log_{10}(test\ value)\) is reported in the output matrix (default). Otherwise the test-value is reported.
3.5 Suffix
The entered suffix will be attached to newly generated columns (default: empty). That way columns from multiple runs of the test can be distinguished more easily.
4 Parameter window