Principal component analysis
1 General
- Type: - Matrix Analysis
- Heading: - Clustering/PCA
- Source code: not public.
2 Brief description
Visualize the projection of the data set defined by PCA in 1, 2 and 3 dimensional viewers.
3 Parameters
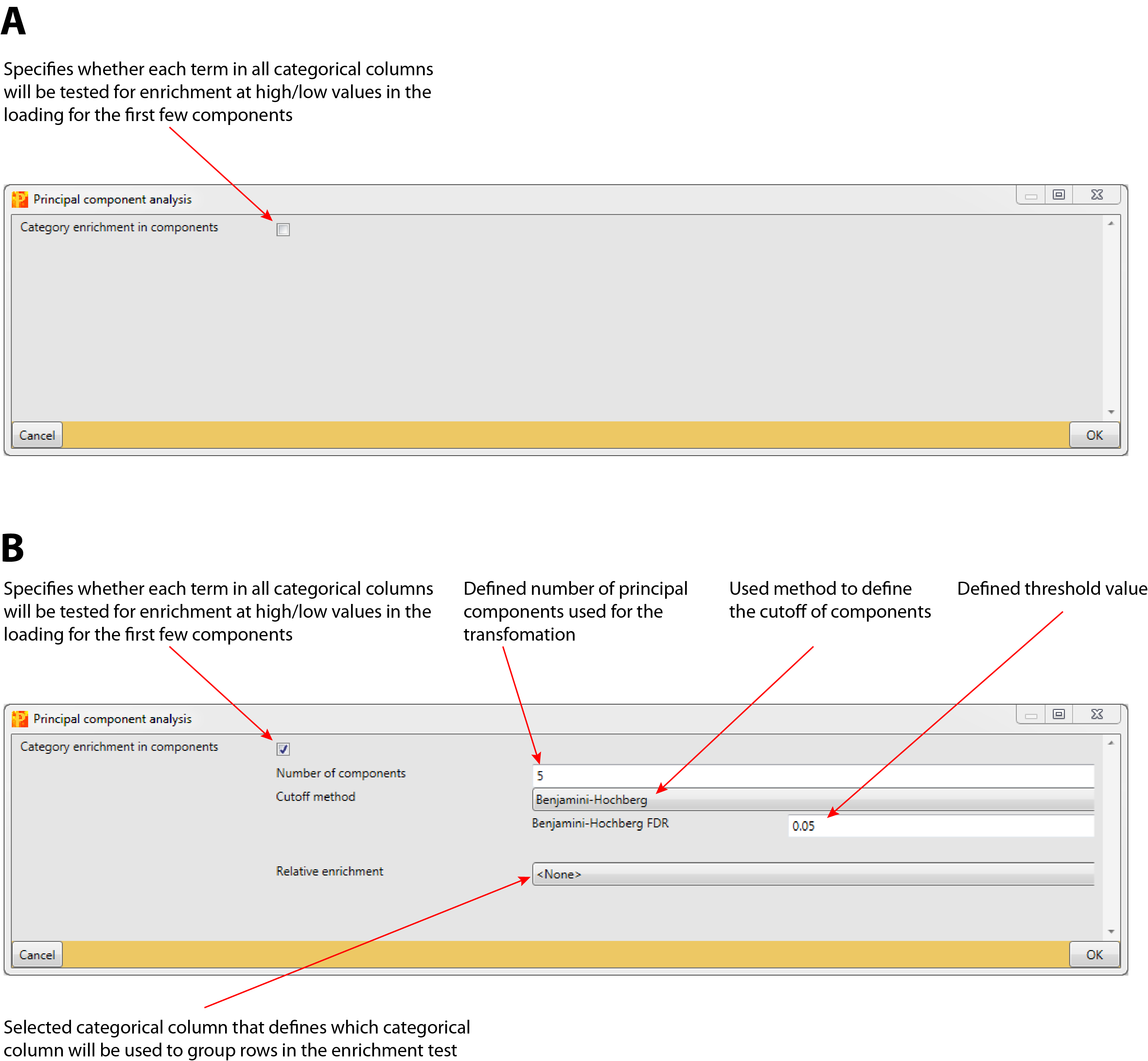
3.1 Category enrichment in components
Specifies, whether each term in all categorical columns will be tested for enrichment at high or low values in the loadings for the first few components (default: unchecked).
3.1.1 Number of components
This parameter is just relevant, if the parameter “Category enrichment in components” is checked. It specifies the number of principal components that will be used for the transformation (default: 5).
3.1.2 Cutoff method
This parameter is just relevant, if the parameter “Category enrichment in components” is checked. It defines the method that is used to calculate the cutoff of potential principal components (default: Benjamini-Hochberg FDR) . The method can be selected from a predefined list:
- Benjamini-Hochberg FDR
- p-value
3.1.3 Threshold
This parameter is just relevant, if the parameter “Category enrichment in components” is checked. It defines the threshold value of the previously specified used cutoff method (default: 0.05).
3.1.4 Relative enrichment
This parameter is just relevant, if the parameter “Category enrichment in components” is checked. It defines, which categorical column will be used to group rows in the enrichment test (default: <None>). All rows having the same identifier will be counted as one entity in the enrichment test. The main application is for posttranslational modification sites. Then one should select here protein or gene identifiers. This will make sure that multiple sites from the same protein (or gene) are counted only once for the enrichment analysis.
4 Parameter window