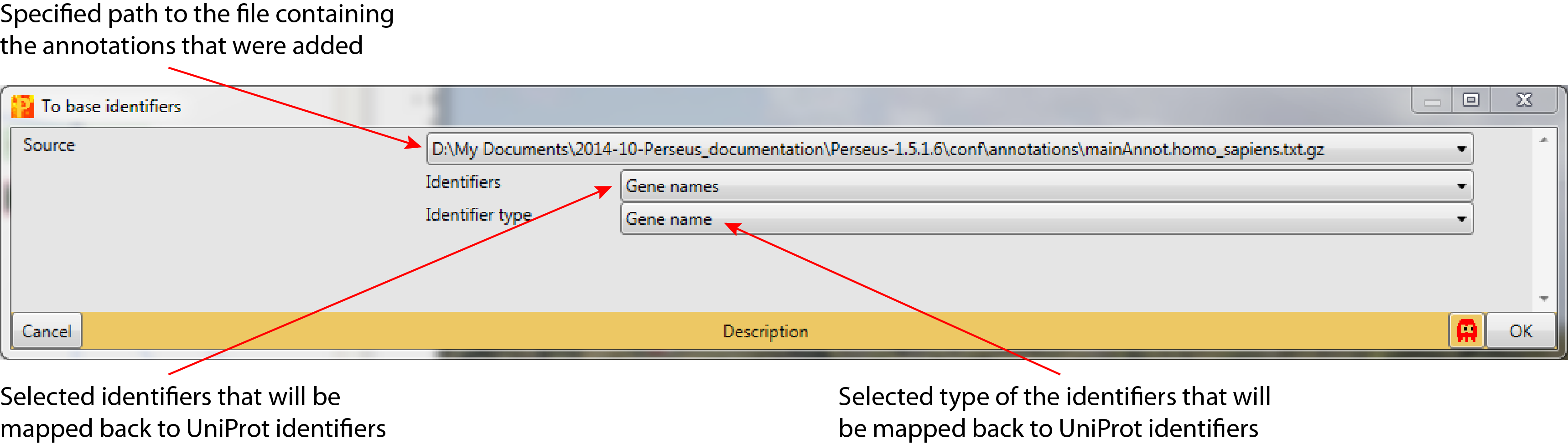
To base identifiers
1 General
- Type: - Matrix Processing
- Heading: - Annot. columns
- Source code: BackToBaseIdentifiers.cs
2 Brief description
This activity does the inverse of the Add annotation activity. Any of the columns that can be created by the Add annotation activity can be mapped back to the base identifiers (typically UniProt ids).
3 Parameters
3.1 Source
Specified path to the file containing the annotations that should be mapped back to the base identifiers (default: first file in Perseus-version /conf/annotations/). The file should be the same that was used to add the annotations, also it can be selected from all files in Perseus-version /conf/annotations/.
3.2 Identifiers
Selected text column containing the identifiers that should be matched back to UniProt identifiers (default: first text column in the matrix).
Comment: Only text columns can be matched back not categorical ones, because there is no unique match.
3.3 Identifier type
Selected type of the identifiers that will be mapped back (default: Gene name). The identifier type can be selected from a predefined list:
- ENSG
- ENSP
- ENST
- Flybase
- Gene name
- MGI
- PDB
- UniProt names
- Wormbase
- EC
- eggNOG
==== Parameter window