Proteomic Ruler
This Perseus plugin implements computational strategies for absolute protein quantification without spike-in standards. The concept was published by Wiśniewski et al.1 and builds on earlier work by Wiśniewski et al.2.
Usually, determining copy numbers of proteins per cell requires the use of spike-in standards in combination with cell counting and accurate protein concentration measurements.
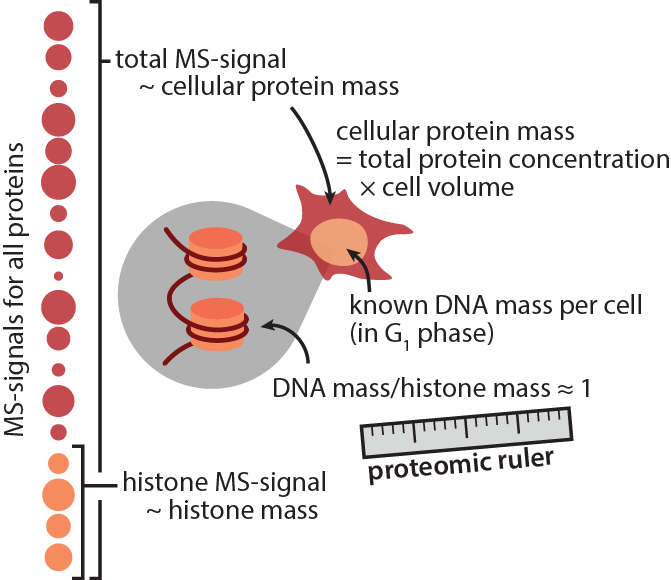
In a nutshell, the idea of the ‘proteomic ruler’ is to use a natural standard instead: Histones. Histones are wrapped around DNA at a defined ratio of ~1:1 (DNA mass/histone mass). The amount of DNA per cell is a function of genome size and ploidy and one usually has good prior knowledge of these factors. Moreover, the total protein concentration is remarkably similar in most cell types, around 200-300 g/l. One can use this to estimate not only copy numbers per cell, but also absolute protein concentrations.
1 Requirements
If you want to use the proteomic ruler plugin on your dataset, you will need a deep, eukaryotic, whole proteome dataset from a nucleated cell type with known ploidy. It is important that you lysed the cells completely and that no cellular fraction was lost during sample preparation. In particular, the chromatin fraction must be included. Reasonably deep proteome coverage is required. From a depth 12,000+ peptides on, we found that the ratio of histone intensity to total intensity flattens out, assuring overall scaling accuracy. However, deeper coverage will be needed to obtain accurate copy numbers of most individual proteins.
If you fractionated your sample, make sure that your fractionation method does not underrepresent histones. We successfully tested SAX, SCX and in-gel fractionation, whereas isoelectric focusing (OFF-gel) fractions tends to underrepresent basic histone peptides, leading to a scaling bias.
For technical reasons, this plugin requires that you searched your data against a recent Uniprot fasta file. The plugin currently supports H. sapiens, M. musculus, D. melanogaster, C. elegans, S. cerevisiae and S. pombe.
2 Installation
As with other Perseus plugins, installation is really easy:
- Download the plugin .dll that is compatible with your Perseus version.
- Copy the .dll into the main Perseus folder.
- Restart Perseus.exe.
- You should then see a new menu item ‘Proteomic ruler’ in the processing section.
3 Usage
Import the proteinGroups.txt of your dataset into Perseus. Make sure to import the following columns:
- Intensity (or LFQ intensity) columns as expression columns
- Peptides, unique, razor + unique peptides as numerical columns
- Protein IDs, Majority protein IDs as text columns
- Sequence length, Molecular weight as numerical columns (optional)
The proteomic ruler plugin comes with three functions that should be executed in this order: